|
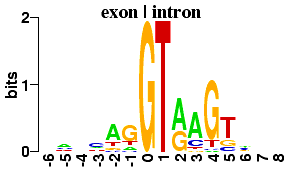
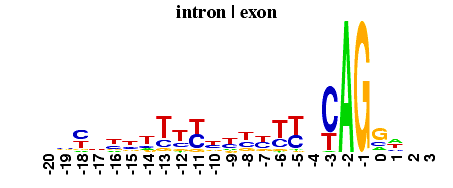
The Edit Logo buttons will transfer the relevant sequence data to the Logo creation form. There you can examine the sequence data and recreate the logo for yourself. Additional examples can be found at the Sequence Logo Gallery.
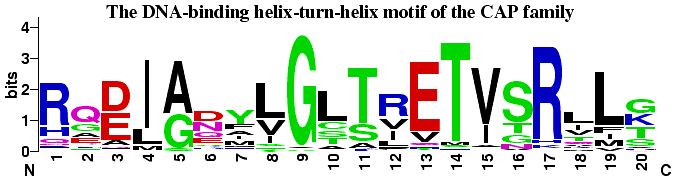 The helix-turn-helix motif from the CAP family of homodimeric DNA
binding proteins. CAP (Catabolite Activator Protein, also known as
CRP for cAMP Receptor Protein) is a transcription promoter that binds
at more than 100 sites within the E. coli genome. Residues 1-7
form the first helix, 8-11 the turn and 12-20 form the DNA recognition
helix. The glycine at position 9 appears to be
critical in forming the turn. Positions 4, 8, 10, 15 and 19 are
partially or completely buried, and therefore tend to be populated by
hydrophobic amino acids, which are colored black. Positions 11-14, 17
and 20 interact directly with bases in the major groove
and are critical to the sequence specific binding of the
protein. The data for this logo consists of 100 sequences from the
full Pfam alignment of this family (Accession number
PF00325). A few sequences with rare insertions were removed for
convenience.
The helix-turn-helix motif from the CAP family of homodimeric DNA
binding proteins. CAP (Catabolite Activator Protein, also known as
CRP for cAMP Receptor Protein) is a transcription promoter that binds
at more than 100 sites within the E. coli genome. Residues 1-7
form the first helix, 8-11 the turn and 12-20 form the DNA recognition
helix. The glycine at position 9 appears to be
critical in forming the turn. Positions 4, 8, 10, 15 and 19 are
partially or completely buried, and therefore tend to be populated by
hydrophobic amino acids, which are colored black. Positions 11-14, 17
and 20 interact directly with bases in the major groove
and are critical to the sequence specific binding of the
protein. The data for this logo consists of 100 sequences from the
full Pfam alignment of this family (Accession number
PF00325). A few sequences with rare insertions were removed for
convenience.
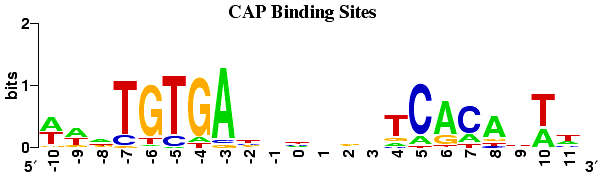 The two DNA recognition helixes of the CAP dimer insert themselves into
into cThe two DNA recognition helixes of the CAP homodimer insert
themselves into consecutive turns of the major groove. Several
consequences can be observed in this CAP binding site logo. The logo
is approximately palindromic, which provides two very similar
recognition sites, one for each subunit of the dimer.
However, the binding
site is not perfectly symmetric, possible due to the
inherent asymmetry of the operon promoter region.
The displacement of the two parts is 11 base pairs, or approximately
one full turn of the DNA helix. Additional interactions between the
protein and the first and last two bases occur within the DNA minor
groove, where it is difficult for the protein to distinguish A from T,
or G from C\cite{Seeman76}.
The data for this logo consists of 59 binding sites determined by
DNA footprinting.
Robison, K., McGuire, A. M., Church, G. M. A comprehensive library of
DNA-binding site matrices for 55 proteins applied to the
complete Escherichia coli K12 genome. Journal of Molecular Biology
(1998) 284, 241-254.
The two DNA recognition helixes of the CAP dimer insert themselves into
into cThe two DNA recognition helixes of the CAP homodimer insert
themselves into consecutive turns of the major groove. Several
consequences can be observed in this CAP binding site logo. The logo
is approximately palindromic, which provides two very similar
recognition sites, one for each subunit of the dimer.
However, the binding
site is not perfectly symmetric, possible due to the
inherent asymmetry of the operon promoter region.
The displacement of the two parts is 11 base pairs, or approximately
one full turn of the DNA helix. Additional interactions between the
protein and the first and last two bases occur within the DNA minor
groove, where it is difficult for the protein to distinguish A from T,
or G from C\cite{Seeman76}.
The data for this logo consists of 59 binding sites determined by
DNA footprinting.
Robison, K., McGuire, A. M., Church, G. M. A comprehensive library of
DNA-binding site matrices for 55 proteins applied to the
complete Escherichia coli K12 genome. Journal of Molecular Biology
(1998) 284, 241-254.
The following logos (along with the CAP logo above) display a selection of E. coli transcription factor binding sites determined by DNA footprinting. This data has been collated in the DPInteract database and has been used to search for additional binding sites within the E. coli genome.
Robison, K., McGuire, A. M., Church, G. M. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K12 genome. Journal of Molecular Biology (1998) 284, 241-254.
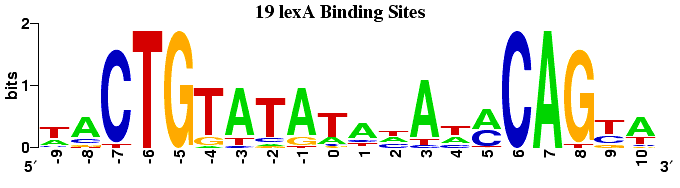
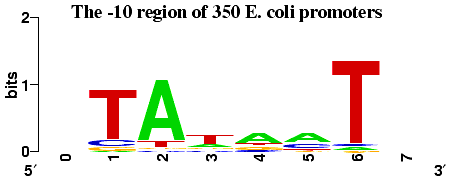
In prokaryotes the DNA sequence just upstream of the transcription start point
contains two important conserved regions. The first such region is centered
at around 35bp upstream and is involved in the initial recognition of the
gene by RNA polymerase.
The second region, sometimes
referred to as the Pribnow box, is centered at about 10bp upstream. The typical
separation between the -35 and -10 sites is 15-18 bp.
See
baseflip:
Strong Minor Groove Base Conservation in Sequence Logos
implies DNA Distortion or Base Flipping during Replication and
Transcription Initiation for more information. This sequence data was kindly provided by Prof. Julia Brettschneider <juliab@stat.berkeley.edu>
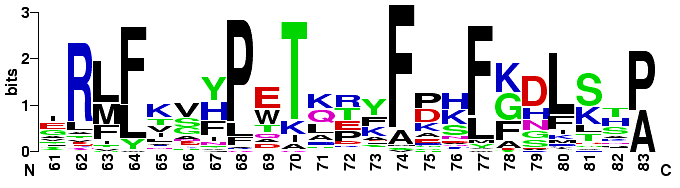 The end of the B helix through the beginning of the D helix of 34 globins. This
sequence data was taken from
Sequence Logos: A New Way to Display Consensus Sequences.
The end of the B helix through the beginning of the D helix of 34 globins. This
sequence data was taken from
Sequence Logos: A New Way to Display Consensus Sequences. Here is an alignment found by the
gibbs
sampling system.
Both the identified site and some context are shown.
Note that spaces are significant, so that
the spaces included below (to aid identification of the site) will end up
being considered amino acid positions.
Here is an alignment found by the
gibbs
sampling system.
Both the identified site and some context are shown.
Note that spaces are significant, so that
the spaces included below (to aid identification of the site) will end up
being considered amino acid positions.
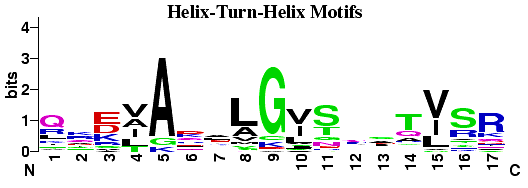 Helix-Turn-Helix DNA binding motifs found by the
gibbs
sampling system. Compared to the CAP HTH logo
there is much less sequence conservation within the DNA binding helix (11-17),
as might be expected for a diverse sample of proteins.
Helix-Turn-Helix DNA binding motifs found by the
gibbs
sampling system. Compared to the CAP HTH logo
there is much less sequence conservation within the DNA binding helix (11-17),
as might be expected for a diverse sample of proteins.